Dynamic bonds
Dynamic bonds is something I invented as an alternative to using complex pair interactions to model DNA hybridization. It is difficult to capture the directionality of a hydrogen bond or a base-pair stacking interaction with a simple distance dependent pair interactions. An alternative is to use anisotropic particles to model the nucleotides, e.g. Guy-Berne or RE-squared interactions. In DNA, it would be nice to have coarse-grained models where the secondary level structure (hybridization) is an explicit binary quantity, and secondly that a nucleotide can bond with exactly one complementary nucleotide. WIth dynamic bonds either there is a hybridization bond or there is no hybridization bond. However, since the force field depends on the instantaneous molecular structure, dynamic bonds belongs to the Hamiltonian replica class of techniques.
I have extended the Large-scale Atomic/Molecular Massively Parallel Simulator (LAMMPS) to support directional bonds and dynamic bonding. The framework supports stochastic formation of new bonds, breakage of existing bonds, and conversion between bond types. Bond formation can be controlled to limit the maximal functionality of a bead with respect to various bond types. Concomitant with the bond dynamics, angular and dihedral interactions are dynamically introduced between newly connected triplets and quartets of beads, where the interaction type is determined from the local pattern of bead and bond types. When breaking bonds, all angular and dihedral interactions involving broken bonds are removed. The framework allows chemical reactions to be modelled, and use it to simulate a simplistic, coarse-grained DNA model.
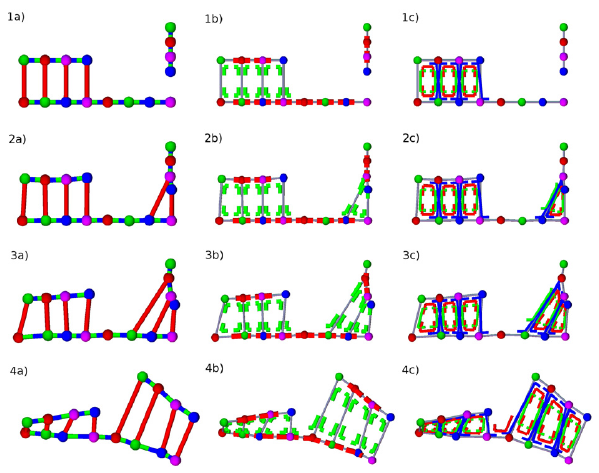
Oligomer – DNA template hybridization (rows 1–4) showing the dynamics of bond, angular, and dihedral interactions (columns a–c) for three times (a,b,c) along which the oligomer becomes hybridized with the template. Note that backbone bond directionality is only shown in the first row for simplicity.
References
